GDSC Single Molecule Light Microscopy (SMLM) ImageJ Plugins¶

GDSC SMLM plugins provide various tools for single molecule localisation analysis.
The tools are easily installed into ImageJ. The library works for Java 1.8+.
The code is hosted on GitHub.
Citation¶

Overview¶
Super-resolution microscopy can be used to obtain a higher level of resolution from an image than conventional light microscopy.
In conventional light microscopy the subject is illuminated and all the light interacting with the subject is captured simultaneously into an image. The resolution is limited since the optics of the microscope cannot focus the light perfectly.
Super-resolution microscopy uses small molecules that can be activated to emit light. If a single molecule is activated its position can be localised using software analysis. Using thousands of localisations it is possible to reconstruct a virtual image of the subject. This increases the image resolution to the precision of the localisation method. The improvement depends on the imaging conditions but is often 5-10 times higher.
Table 1 shows the increased resolution of single molecule localisation microscopy. The difference is most noticeable where structures meet since the single molecules have a cleaner signal when activated individually than the combined signal of all the molecules together.
(A) Standard image
 |
(B) Super-resolution image
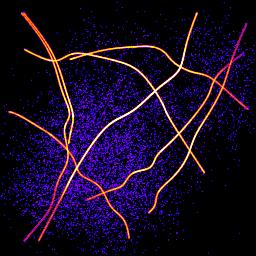 |
(C) Standard image 8x
 |
(D) Super-resolution image 8x
 |
The GDSC Single Molecule Light Microscopy (SMLM) plugins provide various tools for single molecule localisation analysis.
Fit an image, or series of images, using a 2D Guassian Point Spread Function (PSF)
Save results to a table, a file, an image and/or to memory
Trace or cluster localisations through time to identify molecules
Correct drift in long time course images
Estimate fluorophore dark-time and blinking rate
Create localisation density images
Create custom PSFs from calibration images
Create simulated data using a Gaussian or custom PSF with configurable molecule populations and diffusion
Calibrate the gain and read noise of your microscope camera
Estimate noise in an image
Estimate resolution using Fourier Ring Correlation (FRC)
The following plugins are available:
Fitting Plugins¶
Performs fitting on an image to generate a table of molecule localisations and an image reconstruction. Provides a step through guide for initial configuration of microscope parameters |
|
Performs fitting on an image to generate a list of molecule localisations. |
|
Allows management of templates for the fitting configuration. |
|
Allows setting the fitting parameters and saving them to file. |
|
Performs fitting on folder of images to generate a list of molecule localisations. |
|
Identifies candidate maxima in an image. |
|
Identifies candidate maxima in a folder of images. |
|
Shows an interactive view of the candidates that will be identified in an image. |
|
Performs PSF fitting on candidate maxima. |
|
Allows interactive identification of maxima using filters and 2D Gaussian fitting. |
|
Provides analysis of the fail count settings used in the |
|
Allows interactive fitting of clicked spots on an image. |
Results Plugins¶
Allows conversion of the localisation results into different formats, e.g. files or images. |
|
Allows multiple files to be loaded into memory. |
|
Allows multiple results sets to be saved to file. |
|
Displays a summary of the results held in memory. |
|
Removes all results held in memory. |
|
Removes selected results held in memory. |
|
Allows the results sets to be renamed. |
|
Allows the frame number of results to be rebuilt assuming a repeating pattern of data and non-data frames. |
|
Allows results held in memory to be calibrated. |
|
Allows the bounds of the results to be updated. |
|
Allows results held in memory to be converted to different units. |
|
Shows the header information from any support localisation results file format. |
|
Draws an overlay on an image using all the localisations from a results dataset. |
|
Loads localisation data from a delimited text file in a user-specified format. |
|
Saves localisation data to a delimited text file in a user-specified format. |
|
Export traced datasets to file. |
|
Imports track categories from file and assigns them to an existing track dataset. |
|
Filters a list of localisations using signal strength, coordinate shift and PSF width. |
|
Filters a set of localisations using a 2D bounding rectangle. |
|
Filters a set of localisations using any |
|
Filters a list of localisations using a custom filter specified using a text description. Multiple filters can be combined with AND/OR operators. |
|
Filters a set of molecules using various criteria. |
|
Splits a set of localisation results using a 2D mask. |
|
Allows the localisations in a dataset to be translated. |
|
Show an interactive 3D view of the localisations in a dataset. |
|
Calculate the match statistics between two results sets. |
|
Calculate the match statistics between two sets of traced molecules. |
|
Calculate the match between two classifications of the same localisations. |
|
Extracts the fitted spots from an image into a stack ordered by the user-selected score. |
|
Creates a mask of a fission yeast cell (S. pombe). |
|
Creates a 3D mask using three 2D masks to define the shape. |
|
Creates a 3D mask using spheres to model a nucleus of a cell. |
Analysis Plugins¶
Corrects stage drift using: sub-image alignment; fiducial markers within an image; reference stack alignment; or a drift file. |
|
Traces molecules through time using time and distance thresholds (using a type of single-linkage clustering). |
|
Clusters molecules through time using time and distance thresholds (using centroid-linkage clustering). |
|
Traces localisations through time and collates them into traces using a probability model to connect localisations. |
|
Allows the |
|
Trace molecules through consecutive frames. Mean-squared displacement analysis is performed on the traces to calculate a diffusion coefficient. |
|
Allows the |
|
Analyses the track length of traced data. |
|
Analyses the local diffusion of tracks within a window and classifies tracks into sub-populations based on diffusion metrics. |
|
Analyses the residence time of stationary (bound) molecules. |
|
Runs the OPTICS algorithm to perform interactive density-based clustering of localisation data. |
|
Runs the DBSCAN algorithm to perform interactive density-based clustering of localisation data. |
|
Draws collections of localisations with the same ID on an image, for example the output from |
|
Calculates the local density around localisations and displays an image coloured using the density score. |
|
Determines the maximum dark time for a fluorophore from localisation data. |
|
Estimate the blinking rate of fluorophores in a results set. |
|
Performs time correlated photo-activated light microscopy (TC-PALM) analysis. |
|
Saves all localisations paired with their neighbour (if present) to file. |
|
Performs filtering on a set of categorised localisation results and computes match statistics for each filter. |
|
Used to prepare a large set of filters for use in the |
|
Performs filtering on a set of categorised localisation results and computes match statistics for each filter defined in the input file. |
|
Allows analysis of the signal and on/off times for fixed fluorophore spots in an image stack. |
|
This plugin provides a named plugin command for the |
|
Perform crosstalk activation analysis of photo-switchable probes under pulsed light activation. |
|
Perform multi-channel super-resolution imaging by means of photo-switchable probes and pulsed light activation. |
|
Estimates the local density using NxN squares around localisations. |
|
Analyses the resolution of an image using Fourier Ring Correlation. |
|
Estimate the spurious correction factor Q for use in |
PC PALM Plugins¶
Prepare a set of localisations for Pair Correlation analysis. |
|
Produce Pair Correlation curve for a set of localisations selected from a super-resolution image. |
|
Performs spatial analysis to plot the molecule density around each localisation as a function of distance from the localisation. |
|
Saves all the PC-PALM results held in memory to a results folder. |
|
Load all the PC-PALM results from a results folder to memory. |
|
Combine multiple Pair Correlation curves and fit them using models for random or clustered distributions. |
|
Find clusters of molecules using a partial centroid-linkage hierarchical clustering algorithm. |
Model Plugins¶
Extracts the PSF from a Z-stack image of markers, e.g. quantum dots or fluorescent beads. |
|
Computes the drift in a PSF image. |
|
Combines several PSF images into a single average PSF. |
|
Diplays the half-width at half-maxima (HWHM) curve for a PSF image. |
|
Allows management of the cubic spline models of point spread functions (PSFs). |
|
Allows management of 2D Gaussian astigmatism models for a 2D astigmatic PSF. |
|
Creates an image by simulating single molecule localisations using a model of photoactivated diffusing fluorophore complexes. The PSF can be simulated or real using an input PSF image |
|
Creates an image by simulating single molecule localisations at a specified density. |
|
Creates an image by simulating single molecule localisations diffusing in tracks that do not overlap in time. |
|
Creates an image by simulating single molecule localisations in a fixed location. |
|
Fit the image created by |
|
Compute statistics on the accuracy and precision of fitting using different methods. Statistics are only computed for all the localisations that were fit successfully by each method. |
|
Creates a sparse image by simulating zero or one localisation per frame. |
|
Load benchmark data using an open image and a localisations text file. |
|
Filter the image created by |
|
Run a batch of filters through the analysis of |
|
Fits all the candidate spots identified by the |
|
Run different filtering methods on a set of benchmark fitting results outputting performance statistics on the success of the filter. |
|
Run different filter control parameters on a set of benchmark fitting results outputting performance statistics on the success of the filter. |
|
Iterates the |
|
Scores a filter against a set of benchmark fitting results. |
|
Fits candidates in a benchmark localisation image as a single or double spot (doublet) and scores the results. |
|
Filters all the fit results produced by the |
|
Compare methods for classifying spot candidates. |
Calibration Plugins¶
Allows calculation of the Point Spread Function (PSF) size given the microscope imaging parameters. |
|
Estimates the PSF by performing fitting on a sample image. |
|
Opens a folder of images and computes a Mean-Variance test to determine the gain and read noise of the microscope camera. Gain can be calculated for standard or Electron Multiplying (EM) cameras |
|
Analysis a white light image from an EM-CCD camera and fits a model to obtain the bias, EM-gain, read noise and average photons per pixel. |
|
Displays a plot of the probability mass function (PMF) of the expected value of a pixel on an EM-CCD camera given an average number of photons. |
|
Analyse the per pixel offset, variance and gain from a sCMOS camera. |
|
Model the on-chip amplification from an EM-CCD camera, CCD or sCMOS camera. |
|
Model the Fisher information from an EM-CCD camera, CCD or sCMOS camera. |
|
Allows management of per-pixel camera models for the fitting configuration. |
|
Simulate molecule diffusion and fit a graph of mean-squared displacement to determine the diffusion coefficient. |
Tool Plugins¶
In addition to the principle plugins for localisation fitting and analysis there are several utility plugins provided.
Performs smoothing on an image (identical to that performed when identifying maxima). |
|
Switches an image to binary (white/black) to allow quick visualisation of localisations. |
|
Resets a binary image back to the standard display. |
|
Perform filtering to remove hot pixels from an image. |
|
Estimate the noise in an image using various methods. |
|
Estimate the background in a region of an image. |
|
Compute the median of an image, on a per-pixel basis, using a rolling window at set intervals. |
|
Produces a background intensity image and a mask from a sample in vivo image. This can be used to simulate realistic fluorophore distributions. |
|
Allow an image to be added as an overlay with a transparent background. |
|
Convolve an image with a kernel from another image. |
|
Opens a TIFF image as a read-only virtual stack image. Images can be extremely large. |
Toolset Plugins¶
These plugins provide quick access to the various SMLM plugins via ImageJ toolsets and a customisable SMLM Tools window.
Adds an |
|
Display a |
|
Create a configuration file allowing the |
|
Register functions that can be used in the |
All of the plugins can be incorporated into ImageJ macros to allow automation of image analysis workflows.
Contents¶
- 1. Background
- 2. Installation
- 3. ImageJ Plugins Overview
- 4. Fitting Plugins
- 4.1. Simple Fit
- 4.2. Peak Fit
- 4.2.1. Imaging Calibration Parameters
- 4.2.2. Gaussian PSF Parameters
- 4.2.3. Maxima Identification Parameters
- 4.2.4. Fitting Parameters
- 4.2.5. Fit Solvers
- 4.2.6. Multiple Peak Fitting Parameters
- 4.2.7. Filtering Parameters
- 4.2.8. Results Parameters
- 4.2.9. Peak Fit Preview
- 4.2.10. Interactive Results Table
- 4.2.11. Live Image Display
- 4.2.12. Image Examples
- 4.2.13. Running Peak Fit
- 4.2.14. Additional Fitting Options
- 4.2.15. Interlaced Data
- 4.3. Template Manager
- 4.4. Fit Configuration
- 4.5. Peak Fit (Series)
- 4.6. Spot Finder
- 4.7. Spot Finder (Series)
- 4.8. Spot Finder (Preview)
- 4.9. Fit Maxima
- 4.10. Gaussian Fit
- 4.11. Fail Count Manager
- 4.12. Spot Fit Tool
- 5. Results Plugins
- 5.1. Results Manager
- 5.2. Batch Load Results
- 5.3. Batch Save Results
- 5.4. Summarise Results
- 5.5. Clear Memory Results
- 5.6. Clear Memory Results (Multi)
- 5.7. Rename Results
- 5.8. Resequence Results
- 5.9. Calibrate Results
- 5.10. Update Results Bounds
- 5.11. Convert Results
- 5.12. Show Results Header
- 5.13. Overlay Results
- 5.14. Load Localisations
- 5.15. Save Localisations
- 5.16. Trace Exporter
- 5.17. Track Population Importer
- 5.18. Trace Viewer
- 5.19. Filter Results
- 5.20. Crop Results
- 5.21. ROI Crop Results
- 5.22. Free Filter Results
- 5.23. Filter Molecules
- 5.24. Split Results
- 5.25. Translate Results
- 5.26. 3D Results Viewer
- 5.27. Results Match Calculator
- 5.28. Trace Match Calculator
- 5.29. Classification Match Calculator
- 5.30. Spot Inspector
- 5.31. Yeast Mask
- 5.32. Depth Mask
- 5.33. Nucleus Mask
- 6. Analysis Plugins
- 6.1. Drift Calculator
- 6.2. Trace Molecules
- 6.3. Cluster Molecules
- 6.4. Dynamic Trace Molecules
- 6.5. Trace Molecules (Multi)
- 6.6. Trace Diffusion
- 6.7. Trace Diffusion (Multi)
- 6.8. Trace Length Analysis
- 6.9. Track Population Analysis
- 6.10. Residence Time Analysis
- 6.11. OPTICS
- 6.12. DBSCAN
- 6.13. Draw Clusters
- 6.14. Density Image
- 6.15. Dark Time Analysis
- 6.16. Blink Estimator
- 6.17. Time Correlated (TC-PALM) Analysis
- 6.18. Neighbour Analysis
- 6.19. Filter Analysis
- 6.20. Create Filters
- 6.21. Filter Analysis (File)
- 6.22. Spot Analysis
- 6.23. Spot Analysis (Add)
- 6.24. Crosstalk Activation Analysis
- 6.25. Pulse Activation Analysis
- 6.26. Density Estimator
- 6.27. Fourier Image Resolution
- 6.28. FIRE Q Estimation
- 7. PC PALM Plugins
- 8. Model Plugins
- 8.1. Optimisation Overview
- 8.2. PSF Creator
- 8.3. PSF Drift
- 8.4. PSF Combiner
- 8.5. PSF HWHM
- 8.6. Cubic Spline Manager
- 8.7. Astigmatism Model Manager
- 8.8. Create Data
- 8.9. Create Simple Data
- 8.10. Create Track Data
- 8.11. Create Benchmark Data
- 8.12. Fit Benchmark Data
- 8.13. Benchmark Analysis
- 8.14. Create Spot Data
- 8.15. Load Benchmark Data
- 8.16. Filter Spot Data
- 8.17. Filter Spot Data (Batch)
- 8.18. Fit Spot Data
- 8.19. Benchmark Filter Analysis
- 8.20. Benchmark Filter Parameters
- 8.21. Iterate Filter Analysis
- 8.22. Score Filter
- 8.23. Doublet Analysis
- 8.24. Doublet Filter Analysis
- 8.25. Smart Spot Ranking
- 9. Calibration Plugins
- 9.1. PSF Calculator
- 9.2. PSF Estimator
- 9.3. Mean-Variance Test
- 9.4. Mean-Variance Test (EM-CCD)
- 9.5. EM-Gain Analysis
- 9.6. EM-Gain PMF
- 9.7. sCMOS Analysis
- 9.8. Camera Model Analysis
- 9.9. Camera Model Fisher Information Analysis
- 9.10. Camera Model Manager
- 9.11. Diffusion Rate Test
- 10. Tools Plugins
- 11. Toolset Plugins
- 12. Localisation Precision
- 13. Comparison Metrics
- 14. MSD Correction
- 15. References
Index¶
Issues¶
Please file a bug report on GitHub.